Deep learning approaches in development by big players in the tech industry can be used by biologists to extract more information from the images they create.
May 1, 2019
JEF AKST
ABOVE: MODIFIED FROM
© ISTOCK.COM
Six years ago, Steve Finkbeiner of the Gladstone Institutes and the University of California, San Francisco, got a call from Google. He and his colleagues had invented a robotic microscopy system to track single cells over time, amassing more data than they knew what to do with. It was exactly the type of dataset that Google was looking for to apply its deep learning approach, a state-of-the-art form of artificial intelligence (AI).
“We generated enough data to be interesting, is basically what they said,” Finkbeiner recalls of the phone conversation. “They were interested in blue-sky ideas—problems that either humans didn’t think would even be possible or things that a computer could do ten times better or faster.”
Deep learning is really dominant at the moment. It’s really changing the field of image analysis.
—Peter Horvath, Hungarian Academy of Sciences
One application that came to Finkbeiner’s mind was to have a neural network—an algorithm that commonly underlies deep learning approaches (see “A Primer: Artificial Intelligence Versus Neural Networks”)—examine microscopy images and draw from them information that researchers had been unable to visually identify. For example, images of unlabeled cells taken with basic light microscopes do not reveal many details beyond the cell’s overall size and shape. If a researcher is interested in a particular biological process, she will typically use stains or fluorescent tags to look for finer-scale cellular or molecular structures. Maybe, Finkbeiner thought, a computer model could learn to see these fine details that scientists couldn’t when looking at untreated samples.
In April of last year, he and his colleagues published their results: after some training, during which the deep learning networks were shown pairs of labeled and unlabeled images, the models were able to differentiate cell types and identify subcellular structures such as nuclei and neuronal dendrites.1 “The upshot of that article: the answer was, resoundingly, yes. You could take images of unlabeled cells and . . . predict images of labeled and unlabeled structures,” says Finkbeiner. “It’s a way to, almost for free, get a lot more information [on] the cells you’re studying.” The very next month, a second team of researchers similarly showed that a deep learning model could identify stem cell–derived endothelial cells without staining.2 (See “Deep Learning Algorithms Identify Structures in Living Cells” here.)
Such is the power of AI, and deep learning in particular. The approach draws inspiration from the neuron-based architecture of the animal nervous system, with many layers of interconnected nodes that communicate information and learn from experience. From basic research needs, such as cell type identification, to public health and biomedical applications, sophisticated machine learning models are changing how researchers interact with visual data.
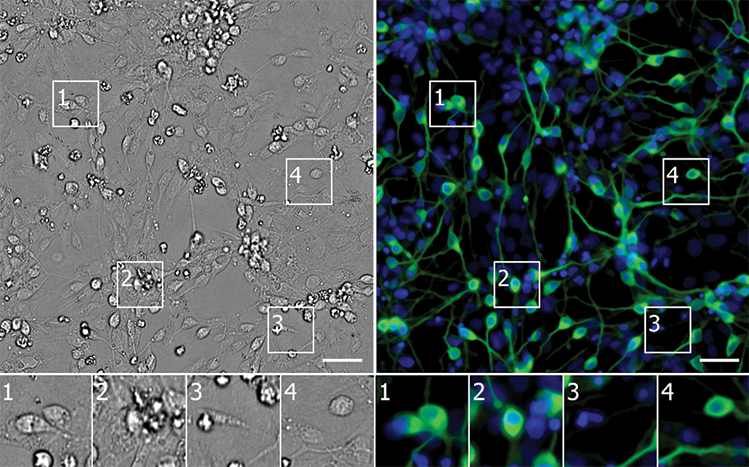
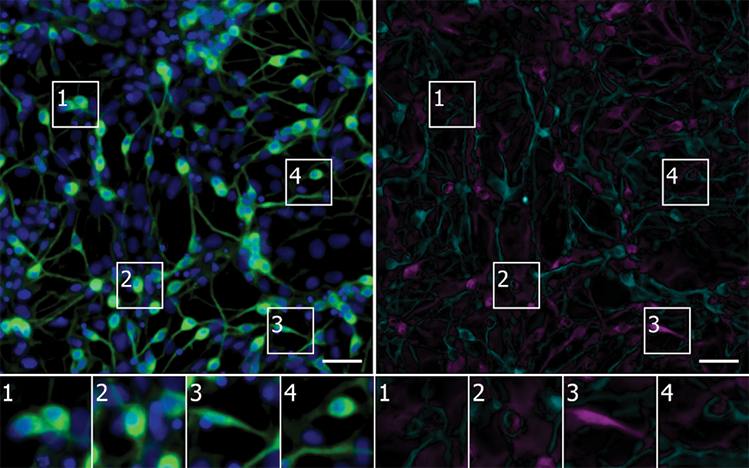 PAINT BY NUMBERS: Using unlabeled images, a neural network identifies neurons derived from induced pluripotent stem cells (green) in a culture containing diverse cell types (nuclei in blue). The error map highlights pixels where the model’s prediction was too bright (magenta) or too dim (teal). Scale bars = 40 mm.
CELL, 173:P792–803.E19, 2018
|
Because of their simplicity and power, these approaches are quickly becoming popular for biological image analyses. “The use of deep learning in microscopy—there was really almost nothing in 2017,” says Samuel Yang, a research scientist at Google who collaborated on the cell microscopy project with Finkbeiner. “In 2018, it [took] off.”
“Deep learning is really dominant at the moment,” agrees Peter Horvath, a computational cell biologist at the Biological Research Centre of the Hungarian Academy of Sciences. “It’s really changing the field of image analysis.”
Identity crisis
There are many ways scientists design intelligent computer systems to identify cells of interest. Last year, Aydogan Ozcan of the University of California, Los Angeles, and colleagues built sensors that focused a deep learning network on microscopy images generated by a mobile phone microscope to screen the air for pollen and fungal spores. The model accurately identified more than 94 percent of samples of the five bioaerosols it was trained on.3 “This is where AI is getting really powerful,” says Ozcan, who notes that traditional screening methods require sending samples out for analysis, while this method can be done on the spot. In 2017, a group of Polish researchers used deep learning algorithms to classify bacterial genera and species in microscopy images, an important part of basic and applied research in fields such as agriculture, food safety, and medicine.4
It’s still pretty early days in the field. But already, we’ve been pretty blown away by how powerful the approach is.
—Steve Finkbeiner, Gladstone Institutes and
University of California, San Francisco
In the biomedical sciences, AI approaches that distinguish among human cell types could improve diagnosis. Finkbeiner has partnered with the Michael J. Fox Foundation for Parkinson’s Research to create a neural network that can differentiate between stem cell–derived neurons from patients with Parkinson’s disease and those from healthy controls. Traditionally, researchers have identified a variable that differs between diseased and healthy cells, but the AI model can learn to use any number of qualities in the image to make its assessment, Finkbeiner says. This multivariate approach should reveal a more complete view of the cell’s health, he says, and help researchers screen for treatments that move a cell toward a healthy state using this more holistic perspective. “With deep learning, you can really transform the way screening is done.”
The use of AI for analysis of microscopy images is nothing new to the cancer field, which has been applying machine learning approaches to analyze micrographs of biopsy samples for more than a decade. In the last couple of years, deep learning has become a popular tool to try to improve diagnoses and treatment regimes. (See “AI Uses Images and Omics to Decode Cancer” here.) After training on images that have been graded by a pathologist, these models can learn to classify various tumors, including lung and ovarian, and to predict cancer progression, often more accurately than clinicians can.
The key to these artificially intelligent models is the computer’s agnostic approach, says Carlos Cordon-Cardo, a physician scientist at the Icahn School of Medicine at Mount Sinai who last year coauthored a study on a machine learning algorithm that graded prostate cancers with a high level of accuracy. “Rather than force the markers, [it’s] more of an open box; the algorithm selects what may be the best players.”

Which elements of the cellular environment are important for a deep learning model to make accurate evaluations is often unclear. “It learns by itself. . . . I cannot say what the neural network does,” says Christophe Zimmer, a computational biophysicist at the Pasteur Institute in Paris. “And the neural network cannot tell me, unfortunately.”
Nonetheless, that information is valuable, as it could provide insights into disease processes that might in turn lead to better diagnostics or treatments. So researchers will “try to break the model,” Finkbeiner explains. By taking away or blurring parts of the image, scientists can see what reduces the model’s performance. This, then, points to those elements as being key for the model’s assessments, yielding clues about what the models “see” in the images that might have some biological underpinning.
“It’s one thing to produce data but another to produce knowledge,” says Cordon-Cardo.
AI improves resolution
Using deep learning to analyze microscopy images is just one example of how AI can influence visual data, says Ozcan. “This is the first wave—let there be an image captured as before . . .and we’ll act on [it] better than before. The second wave is even more exciting to me. That is how AI is helping us with image generation.”
Finkbeiner’s work with Google is one example. The deep neural network can, using brightfield images, generate images that look like fluorescence micrographs. And Ozcan and his colleagues have used deep learning approaches to improve the quality of smartphone microscopes5 and create holographic reconstructions to visualize objects in three dimensions based on a single snapshot.6 Most recently, his group enhanced basic images taken with benchtop microscopes into images of a quality similar to or better than those taken by Nobel Prize–winning super-resolution technologies.7 (See “AI Networks Generate Super-Resolution from Basic Microscopy,” The Scientist, December 17, 2018.) These improved images can then be fed into deep learning models that analyze their content, Ozcan says.
  UPSAMPLING: Deep learning approaches can transform images taken with a smartphone microscope into an image akin to a micrograph taken with a benchtop instrument (bottom), or turn such standard micrographs into super-resolution captures (top).
OZCAN LAB AT UCLA
|
“It’s still pretty early days in the field,” says Finkbeiner. But already, “we’ve been pretty blown away by how powerful the approach is.” His team is now adapting its robotic system to integrate AI directly into microscopes, teaching them to recognize individual cells not by their physical location but by “facial” recognition. One day, he says, researchers may be able to program the system to conduct experiments autonomously—delivering drugs to certain cells based on what it observes, for example—which “could accelerate discovery and lead to less bias,” Finkbeiner says. (See “Robert Murphy Bets Self-Driving Instruments Will Crack Biology’s Mysteries” here.)
In addition to being powerful, AI paired with microscopy is user-friendly. “Deep learning is just so powerful and so easy to use just kind of out of the box,” says Yang, if you have the right problem and data. He says he was surprised the first time a colleague showed him that a deep neural network model trained on images of dogs, cats, and cars from the internet could be adapted to cluster microscopy images of breast cancer cells “almost perfectly” based on what type of drug the cells had been treated with. For cellular applications, most researchers will recommend their models be trained on relevant sets of images to ensure accuracy, but experts say the approach itself is essentially transferrable.
“If there is a state-of-the-art paper in computer vision, it doesn’t really take any modification for it to be applicable to biology,” says Allen Goodman, a computer scientist at the Broad Institute of MIT and Harvard. “It’s Google, Apple, etc.—we can benefit from these methods almost immediately.”
References
- E.M. Christiansen et al., “In silico labeling: Predicting fluorescent labels in unlabeled images,” Cell, 173:P792–803.E19, 2018.
- D. Kusumoto et al., “Automated deep learning–based system to identify endothelial cells derived from induced pluripotent stem cells,” Stem Cell Rep, 10:P1687–95, 2018.
- Y. Wu et al., “Label-free bioaerosol sensing using mobile microscopy and deep learning,” ACS Photon, 5:4617–27, 2018.
- B. Zieli?ski et al., “Deep learning approach to bacterial colony classification,” PLOS ONE, 12:e0184554, 2017.
- Y. Rivenson et al., “Deep learning enhanced mobile-phone microscopy,” ACS Photon, 5:2354–64, 2018.
- Y. Rivenson et al., “Phase recovery and holographic image reconstruction using deep learning in neural networks,” Light-Sci Appl, 7:17141, 2018.
- H. Wang et al., “Deep learning enables cross-modality super-resolution in fluorescence microscopy,” Nat Methods, 16:103–10, 2019.


