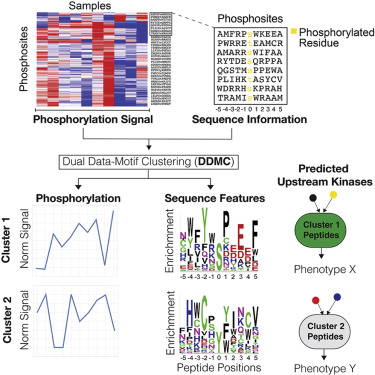
Cancer cells respond to their environment through the activity of signaling proteins called kinases. These proteins are especially important because several cancer therapies target hyperactive kinases that drive tumor growth and spread. Creixell and Meyer built a computational algorithm that characterizes the overall signaling changes in cancer cells to identify drug targets. They tested the utility of their method by analyzing over 100 lung cancer tumors and normal tissue samples from the same patients. This analysis identified kinase activity patterns that are consistently changed in cancer, are associated with specific cancer mutations, and are related to anti-cancer immune responses. This algorithm, therefore, provides a better way to identify kinases that may be effective drug targets.